Econometric models
Probit and logit models
In the probit and logit models dependent variable is dummy variable (0 and 1). For example, whether you defaulted on your credit or not. Whether you are eligible for the program or not, etc etc.
Data source
For more details please refere to AER package documentation, page 140. link Cross-section data about resume, call-back and employer information for 4,870 fictitious resumes sent in response to employment advertisements in Chicago and Boston in 2001, in a randomized controlled experiment conducted by Bertrand and Mullainathan (2004). The resumes contained information concerning the ethnicity of the applicant. Because ethnicity is not typically included on a resume, resumes were differentiated on the basis of so-called "Caucasian sounding names" (such as Emily Walsh or Gregory Baker) and "African American sounding names" (such as Lakisha Washington or Jamal Jones). A large collection of fictitious resumes were created and the presupposed ethnicity (based on the sound of the name) was randomly assigned to each resume. These resumes were sent to prospective employers to see which resumes generated a phone call from the prospective employer.
Logit model
library(AER)
data(ResumeNames)
head(ResumeNames)
| name | gender | ethnicity | quality | call | city | jobs | experience | honors | volunteer | ... | minimum | equal | wanted | requirements | reqexp | reqcomm | reqeduc | reqcomp | reqorg | industry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | <int> | <int> | <fct> | <fct> | ... | <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | <fct> | |
| 1 | Allison | female | cauc | low | no | chicago | 2 | 6 | no | no | ... | 5 | yes | supervisor | yes | yes | no | no | yes | no | manufacturing |
| 2 | Kristen | female | cauc | high | no | chicago | 3 | 6 | no | yes | ... | 5 | yes | supervisor | yes | yes | no | no | yes | no | manufacturing |
| 3 | Lakisha | female | afam | low | no | chicago | 1 | 6 | no | no | ... | 5 | yes | supervisor | yes | yes | no | no | yes | no | manufacturing |
| 4 | Latonya | female | afam | high | no | chicago | 4 | 6 | no | yes | ... | 5 | yes | supervisor | yes | yes | no | no | yes | no | manufacturing |
| 5 | Carrie | female | cauc | high | no | chicago | 3 | 22 | no | no | ... | some | yes | secretary | yes | yes | no | no | yes | yes | health/education/social services |
| 6 | Jay | male | cauc | low | no | chicago | 2 | 6 | yes | no | ... | none | yes | other | no | no | no | no | no | no | trade |
# encoding "call" column categorical levels into numerical
ResumeNames$call <- ifelse(ResumeNames$call == "yes", 1, 0)
# making logit model
model_logit <- glm(call ~ethnicity + gender + quality, data = ResumeNames)
summary(model_logit)
Call:
Call:
glm(formula = call ~ ethnicity + gender + quality, family = binomial(link = "logit"),
data = ResumeNames)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.4770 -0.4356 -0.3867 -0.3525 2.4207
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.4350 0.1348 -18.058 < 2e-16 ***
ethnicityafam -0.4399 0.1074 -4.096 4.2e-05 ***
genderfemale 0.1276 0.1289 0.990 0.3222
qualityhigh 0.1914 0.1059 1.806 0.0709 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 2726.9 on 4869 degrees of freedom
Residual deviance: 2705.7 on 4866 degrees of freedom
AIC: 2713.7
Number of Fisher Scoring iterations: 5
From the regression table we can see coefficient for ethnicityafam is -0.4399, that means that if the applicant have african american sounding name then he is less likely to recieve call back.
We can not interpret magnitude from the regression table for logit model, only we can interpret the direction of the effect i.e. more likely or less likely get called back.
Probit model
Probit models are pretty much similiar to logit models(see above). Just in the glm() command we need to specify the family argument to be family = binomial(link="logit").
model_probit <- glm(call ~ ethnicity + gender + quality, family = binomial (link="probit"), data = ResumeNames)
summary(model_probit)
Call:
glm(formula = call ~ ethnicity + gender + quality, family = binomial(link = "probit"),
data = ResumeNames)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.4757 -0.4363 -0.3871 -0.3525 2.4225
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.39798 0.06642 -21.047 < 2e-16 ***
ethnicityafam -0.21685 0.05282 -4.105 4.04e-05 ***
genderfemale 0.06208 0.06336 0.980 0.3272
qualityhigh 0.09305 0.05252 1.772 0.0764 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 2726.9 on 4869 degrees of freedom
Residual deviance: 2705.8 on 4866 degrees of freedom
AIC: 2713.8
Number of Fisher Scoring iterations: 5
From the regression table we can see coefficient for ethnicityafam is -0.21685 (which is little bit different compared to logit model) that means that if the applicant have african american sounding name then he is less likely to recieve call back.
We can not interpret magnitude from the regression table for probit model, only we can interpret the direction of the effect i.e. more likely or less likely get called back.
Average marginal effects
To have meaningful interpretation of effects we can calculate average marginal effects.
Logit model
marginal_logit <- mean(dlogis(predict(model_logit, type="link")))
marginal_logit * coef(model_logit)
(Intercept) -0.179439881066858 ethnicityafam -0.324139363996093 genderfemale 0.00940104037935758 qualityhigh 0.0141013274125201
Interpretation of marginal effect for variable ethnicityafam is -0.324139363996093, which means if the person has african american sounding name, then he is 32% less likely to get call back from potential employer.
Probit model
marginal_probit <- mean(dnorm(predict(model_probit, type="link")))
marginal_probit * coef(model_probit)
(Intercept) -0.207308146098111 ethnicityafam -0.0321574328415445 genderfemale 0.00920546735038312 qualityhigh 0.0137988149880605
Interpretation of marginal effect for variable ethnicityafam is -0.0321574328415445, which means if the person has african american sounding name, then he is 32% less likely to get call back from potential employer.
We can see that coefficient for logit and probit models could be quite different, but the average marginal effects are on contrary quite similliar.
Multinomial logit model
If the outcome variable is categorical variable without inherent oreder(regular categorical), such as car manufacturers. The we need to use multinomial logit model.
Data source
Data is on penguins and their characterstics. There is three type of penguins: Adelie, Gentoo and Chinstrap. Data were collected and made available by Dr. Kristen Gorman and the Palmer Station, Antarctica LTER, a member of the Long Term Ecological Research Network.
library(nnet)
df <- read.csv('https://raw.githubusercontent.com/mwaskom/seaborn-data/master/penguins.csv')
head(df)
multinom_logit <- multinom(species ~ bill_length_mm + flipper_length_mm + body_mass_g + sex, data = df)
summary(multinom_logit)
Call:
multinom(formula = species ~ bill_length_mm + flipper_length_mm +
body_mass_g + sex, data = df)
Coefficients:
(Intercept) bill_length_mm flipper_length_mm body_mass_g sexFEMALE sexMale
Chinstrap -126.8121 3.884542 0.05522745 -0.013713867 1.140113 -9.134096
Gentoo -190.0349 1.235787 0.59656936 0.005336206 -2.307075 -17.078332
Std. Errors:
(Intercept) bill_length_mm flipper_length_mm body_mass_g sexFEMALE sexMALE
Chinstrap 0.031418912 1.0980540 0.1551344 0.006644753 0.06812109 0.03713893
Gentoo 0.007206881 0.9901455 0.2358709 0.006981574 0.08631697 0.03341686
Residual Deviance: 7.341373
AIC: 31.34137
These are the logit coefficients relative to the reference category. For example, under ‘body_mass_g', the 0.006644753 suggests that for one unit increase in ‘body_mass_g' weight, the logit coefficient for ‘Chinstrap' relative to ‘Adelie' will go up by that amount, 0.006644753. In other words, if your ‘body_mass_g' weight increases one unit, the chances of the penguin to be identified as ‘Chinstrap' compared to the chances of being identified as ‘Adelie' are higher.
Multinom() function does not provide p-values. Either you can compute them customly, or you can use package stargazer, that computes p-values for you.
library(stargazer)
stargazer(multinom_logit, type="text")
==============================================
Dependent variable:
----------------------------
Chinstrap Gentoo
(1) (2)
----------------------------------------------
bill_length_mm 3.885*** 1.236
(1.098) (0.990)
flipper_length_mm 0.055 0.597**
(0.155) (0.236)
body_mass_g -0.014** 0.005
(0.007) (0.007)
sexFEMALE 1.140*** -2.307***
(0.068) (0.086)
sexMALE -9.134*** -17.078***
(0.037) (0.033)
Constant -126.812*** -190.035***
(0.031) (0.007)
----------------------------------------------
Akaike Inf. Crit. 31.341 31.341
==============================================
Note: *p<0.1; **p<0.05; ***p<0.01
Relative risk ratios
Relative risk ratios allow an easier interpretation of the logit coefficients. They are the exponentiated value of the logit coefficients.
multinom_logit_coeff <- exp(coef(multinom_logit))
stargazer(multinom_logit, type="text", coef=list(multinom_logit_coeff))
==============================================
Dependent variable:
----------------------------
Chinstrap Gentoo
(1) (2)
----------------------------------------------
bill_length_mm 48.645 3.441
(1.098) (0.990)
flipper_length_mm 1.057*** 1.816***
(0.155) (0.236)
body_mass_g 0.986*** 1.005***
(0.007) (0.007)
sexFEMALE 3.127*** 0.100***
(0.068) (0.086)
sexMALE 0.0001*** 0.00000***
(0.037) (0.033)
Constant 0.000 0.000
(0.031) (0.007)
----------------------------------------------
Akaike Inf. Crit. 31.341 31.341
==============================================
Note: *p<0.1; **p<0.05; ***p<0.01
- If we look at the first row of the regression table, we can interpret it as following:
- keeping all other variables constant, if bill_length_mm varaible increases one unit(cm), you are 48.645 times more likely to be identified as Chinstrap penguin as compared to the Adelie penguins. The coefficient, however, is not significant.
- Keeping all other variables constant, if bill_length_mm varaible increases one unit(cm), you are 3.441 times more likely to be identified as Gentoo penguin as compared to the Adelie penguins. The coefficient, however, is not significant.
Mutlinomial probit model
Polynomial regression
In some cases, the true relationship between the outcome and a predictor variable might not be linear.
There are different solutions extending the linear regression model for capturing these nonlinear effects, including:
-
Polynomial regression. This is the simple approach to model non-linear relationships. It add polynomial terms or quadratic terms (square, cubes, etc) to a regression.
-
Spline regression. Fits a smooth curve with a series of polynomial segments. The values delimiting the spline segments are called Knots.
-
Generalized additive models (GAM). Fits spline models with automated selection of knots.
We'll use Boston data set. The median house value (mdev), in Boston Suburbs. Variable lstat (percentage of lower status of the population).
library(tidyverse)
data("Boston", package = "MASS")
head(Boston)
| crim | zn | indus | chas | nox | rm | age | dis | rad | tax | ptratio | black | lstat | medv | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 0.00632 | 18 | 2.31 | 0 | 0.538 | 6.575 | 65.2 | 4.0900 | 1 | 296 | 15.3 | 396.90 | 4.98 | 24.0 |
| 2 | 0.02731 | 0 | 7.07 | 0 | 0.469 | 6.421 | 78.9 | 4.9671 | 2 | 242 | 17.8 | 396.90 | 9.14 | 21.6 |
| 3 | 0.02729 | 0 | 7.07 | 0 | 0.469 | 7.185 | 61.1 | 4.9671 | 2 | 242 | 17.8 | 392.83 | 4.03 | 34.7 |
| 4 | 0.03237 | 0 | 2.18 | 0 | 0.458 | 6.998 | 45.8 | 6.0622 | 3 | 222 | 18.7 | 394.63 | 2.94 | 33.4 |
| 5 | 0.06905 | 0 | 2.18 | 0 | 0.458 | 7.147 | 54.2 | 6.0622 | 3 | 222 | 18.7 | 396.90 | 5.33 | 36.2 |
| 6 | 0.02985 | 0 | 2.18 | 0 | 0.458 | 6.430 | 58.7 | 6.0622 | 3 | 222 | 18.7 | 394.12 | 5.21 | 28.7 |
ggplot(Boston, aes(lstat, medv) ) +
geom_point() +
geom_smooth(method='lm')
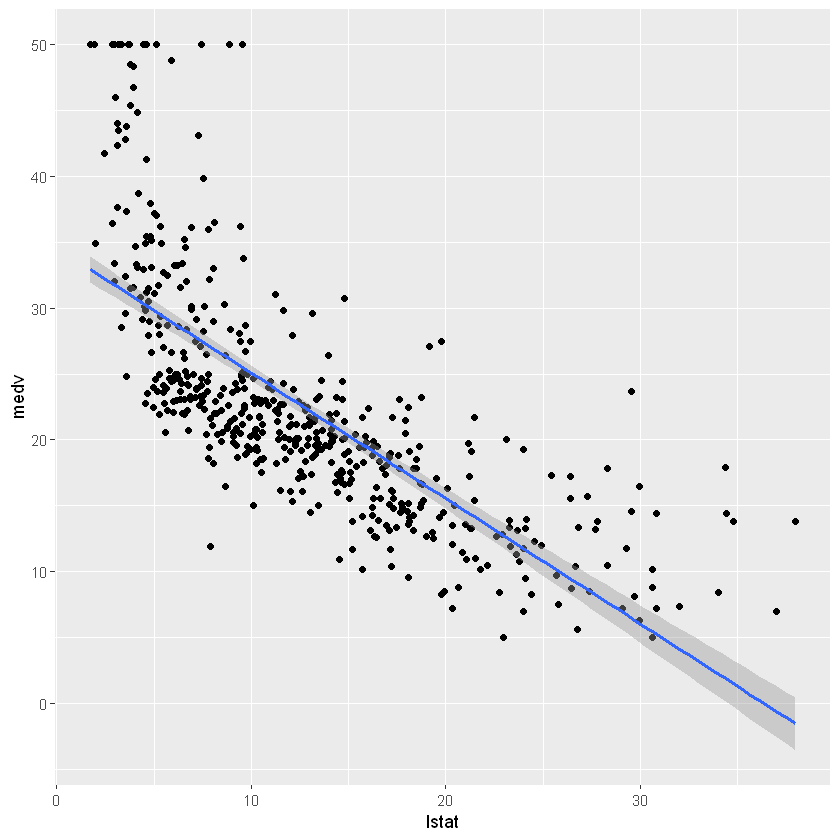
model <- lm(medv ~ lstat, data = Boston)
summary(model)
Call:
lm(formula = medv ~ lstat, data = Boston)
Residuals:
Min 1Q Median 3Q Max
-15.168 -3.990 -1.318 2.034 24.500
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 34.55384 0.56263 61.41 <2e-16 ***
lstat -0.95005 0.03873 -24.53 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.216 on 504 degrees of freedom
Multiple R-squared: 0.5441, Adjusted R-squared: 0.5432
F-statistic: 601.6 on 1 and 504 DF, p-value: < 2.2e-16
The polynomial regression adds polynomial or quadratic terms to the regression equation as follow:
\[medv = b0 + b1*lstat + b2*lstat^2\]model_pol <- lm(medv ~ lstat + I(lstat^2), data = Boston)
summary(model_pol)
Call:
lm(formula = medv ~ lstat + I(lstat^2), data = Boston)
Residuals:
Min 1Q Median 3Q Max
-15.2834 -3.8313 -0.5295 2.3095 25.4148
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 42.862007 0.872084 49.15 <2e-16 ***
lstat -2.332821 0.123803 -18.84 <2e-16 ***
I(lstat^2) 0.043547 0.003745 11.63 <2e-16 ***
---
Signif. codes: 0 ‘***' 0.001 ‘**' 0.01 ‘*' 0.05 ‘.' 0.1 ‘ ' 1
Residual standard error: 5.524 on 503 degrees of freedom
Multiple R-squared: 0.6407, Adjusted R-squared: 0.6393
F-statistic: 448.5 on 2 and 503 DF, p-value: < 2.2e-16
model_pol6 <- lm(medv ~ poly(lstat, 6, raw = TRUE), data = Boston)
summary(model_pol6)
Call:
lm(formula = medv ~ poly(lstat, 6, raw = TRUE), data = Boston)
Residuals:
Min 1Q Median 3Q Max
-14.7317 -3.1571 -0.6941 2.0756 26.8994
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.304e+01 5.593e+00 13.059 < 2e-16 ***
poly(lstat, 6, raw = TRUE)1 -1.517e+01 2.965e+00 -5.115 4.49e-07 ***
poly(lstat, 6, raw = TRUE)2 1.930e+00 5.713e-01 3.378 0.000788 ***
poly(lstat, 6, raw = TRUE)3 -1.307e-01 5.202e-02 -2.513 0.012295 *
poly(lstat, 6, raw = TRUE)4 4.686e-03 2.407e-03 1.947 0.052066 .
poly(lstat, 6, raw = TRUE)5 -8.416e-05 5.450e-05 -1.544 0.123186
poly(lstat, 6, raw = TRUE)6 5.974e-07 4.783e-07 1.249 0.212313
---
Signif. codes: 0 ‘***' 0.001 ‘**' 0.01 ‘*' 0.05 ‘.' 0.1 ‘ ' 1
Residual standard error: 5.212 on 499 degrees of freedom
Multiple R-squared: 0.6827, Adjusted R-squared: 0.6789
F-statistic: 178.9 on 6 and 499 DF, p-value: < 2.2e-16
ggplot(Boston, aes(lstat, medv) ) +
geom_point() +
geom_smooth(method = lm, formula = y ~ poly(x, 4, raw = TRUE))
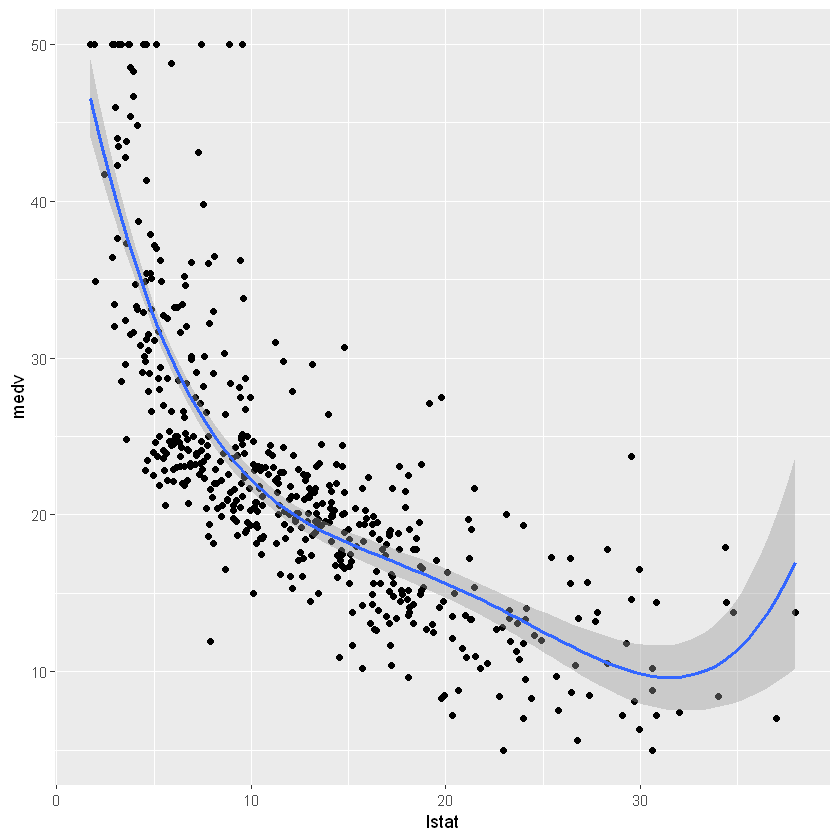
Quantile regression
Quantile regression is an extension of linear regression used when the conditions of linear regression are not met. One advantage of quantile regression relative to ordinary least squares regression is that the quantile regression estimates are more robust against outliers in the response measurements.
For quantile regression to be feasible, dependent variable should have not many zeros or repeated values